Traing Data for Neuron Reconstructors
January 27, 2020
The Cell Types Database is one of the major data products of The Allen Institute for Brain Science. In the Cell Types project, the Allen is constructing an altas of all type of cells found in brains of mice and humans.
There are multiple ways different cells are represented in the Database: electrophysiology spike train recordings, simulation models (GLIF or perisomatic), etc. Of particular interest for the Reconstrue project is the morphology data – the skeletons in the *.swc files.
The Allen has created about 500 SWC files for mouse neurons. In the following Venn diagram of all The Allen’s mouse cells, those ~500 SWC files are grouped inside the red circle.

The main problem from The Allen’s perspective is that they would like to have the red circle be as big as the main outer circle. Each SWC files represents many hours of manual labor by trained specialists reviewing and editing the SWC file. The Allen processes hundreds of such cells a year. Creating skeleton reconstructions is currently a serious manual labor bottleneck.
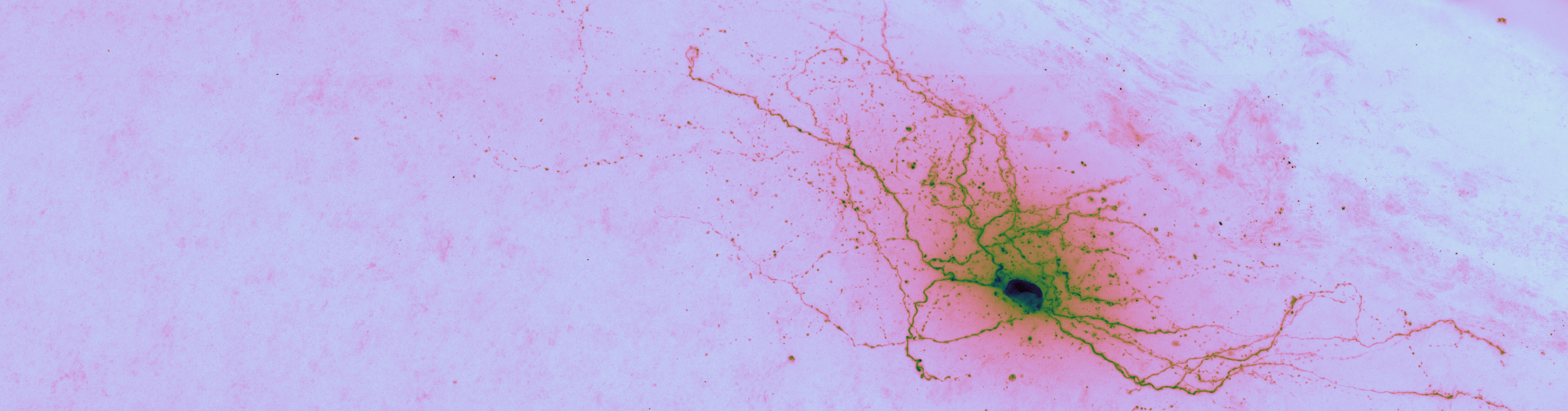
The header image of this post is an example brightfield imaged neuron from The Cell Types Database. This would seem like the sort of object recognition that CNNs and friends (RNNs, FFNs, U-Net, etc.) could automate. This is proving to be nontrivial.
Model training data
The image stack is the input to the machine to be built, and the SWC file is the output.
From a model training perspective, the skeleton in an SWC file can be seen as the “labels” (labeling each voxel in a volume as either inside or outside the cell) for “the training data” (read: the brightfield image stacks). As such, for the purpose of training reconstruction models, we’re only interested in the subset of cells in the atlas Cell Types Database that have skeletons and a microscopy image stack.
!pip install --quiet allensdk# Query the Cell Types DB for files with skeletons a.k.a. reconstructions
# via https://allensdk.readthedocs.io/en/latest/cell_types.html#cell-types-cache
from allensdk.core.cell_types_cache import CellTypesCache
ctc = CellTypesCache(manifest_file='cell_types/manifest.json')
# a list of cell metadata for cells with reconstructions, download if necessary
cells = ctc.get_cells(require_reconstruction=True)
print('Number of cells with SWC files: %i' % len(cells))Number of cells with SWC files: 637Some of those are human cells, atop the roughly 500 mouse cells. Humans brains are much bigger than mouse brains. Training should focus on one species. The Allen has many more mouse neurons than human neurons. So, train on mouse neurons only.
from allensdk.api.queries.cell_types_api import CellTypesApi
# We want mouse cells that have images and skeletons, both.
# Former is data; latter is training labels a.k.a. gold standards.
cells = ctc.get_cells(require_reconstruction=True, require_morphology=True, species=[CellTypesApi.MOUSE])
print('Number of mouse cells with images and SWC files: %i' % len(cells))Number of mouse cells with images and SWC files: 485So, for brightfield reconstructor training, The Allen’s Cell Types Database can be used as a labeled training dataset consisting of about 500 samples. That’s somewhere on the order of ten petabytes of training data.