Brightfield Auto-Reconstruction Challenge
August 25, 2019
Abstract
For the BioImage Informatics 2019 Conference, the Allen Institute issued a Brightfield Auto-Reconstruction Challenge. Neuron object recognition in brightfield microscopy is an open problem, currently necessitating many hours of manual neurite tracing. The Challenge’s ~2.5 terabyte dataset is available for use in research on brightfield microscopy neuron reconstruction.
This project is a collection of Jupyter notebooks that perform data analysis on the Challenge dataset. These notebooks were developed on and are hosted on Google Colab and so can be re-run by anyone wishing to reproduce the same analysis, without any software tooling set-up.

Introduction
For the Brightfield Auto-Reconstruction Challenge, the Allen Institute assembled a dataset containing 115 cells for use in an evalution competition. The dataset is about 2.5 terabytes in size, consisting of raw brightfield microscopy image stacks produced by the Allen Institute.
The dataset also includes SWC files for 105 of those 115 cells. These SWCs contain neuron skeletons manually traced by human experts. These stick figures are the labels for the trainging data – the so called gold standards i.e. the best answers as manually generated by human experts involving many hours of tedious data entry a.k.a neurite tracing.
That works out to a roughly 90%/10% split of the datase into train and test subsets. The challenge is to generate SWCs for the ten cells in the test set.
This project presents the code for multiple methods of neuron reconstruction including ShuTu and U-Net, two methods used in the original Challenge at BioImage 2019. Current research is exploring ResNet and Flood-filling Networks (FFNs) techniques.
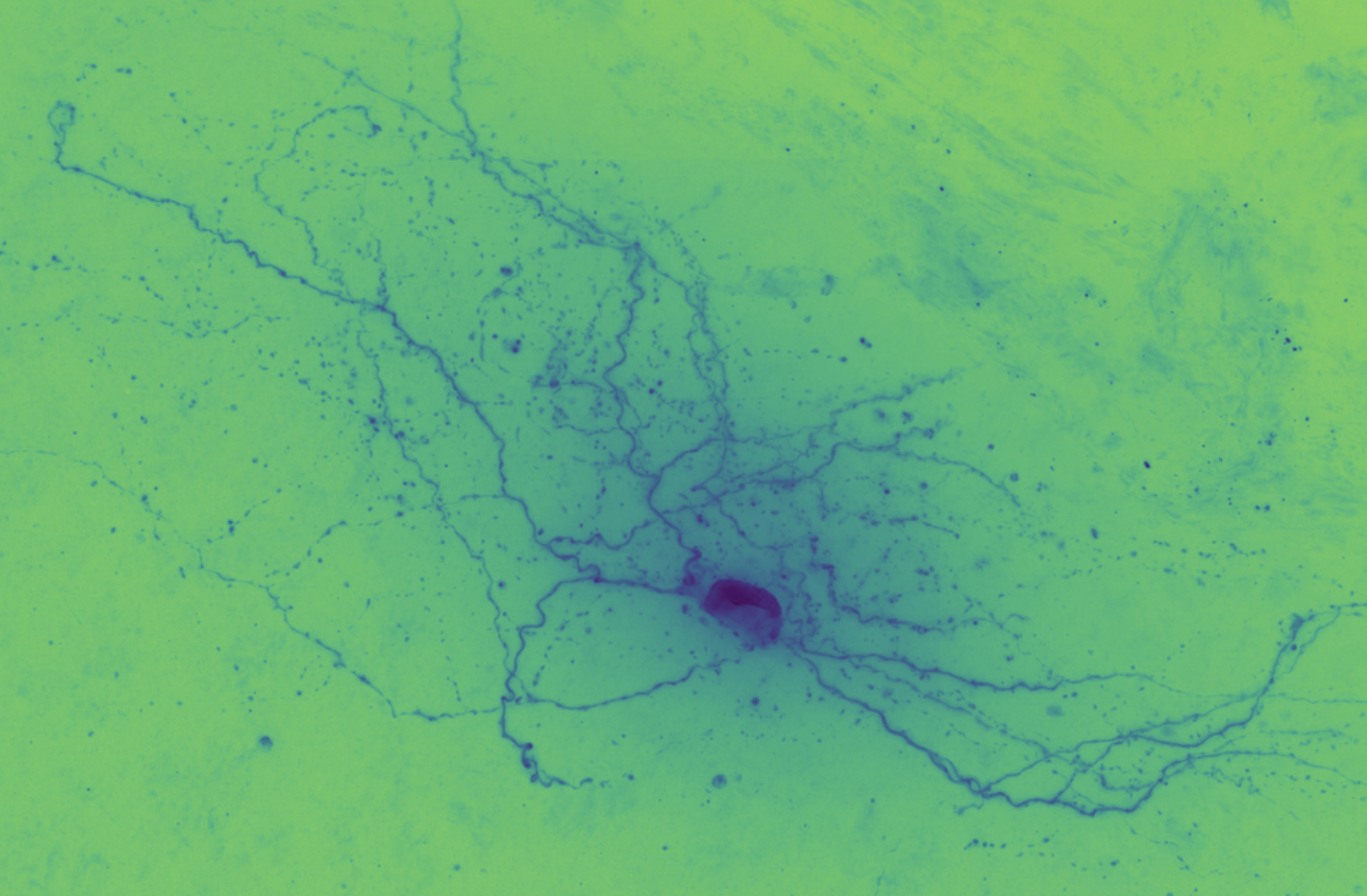
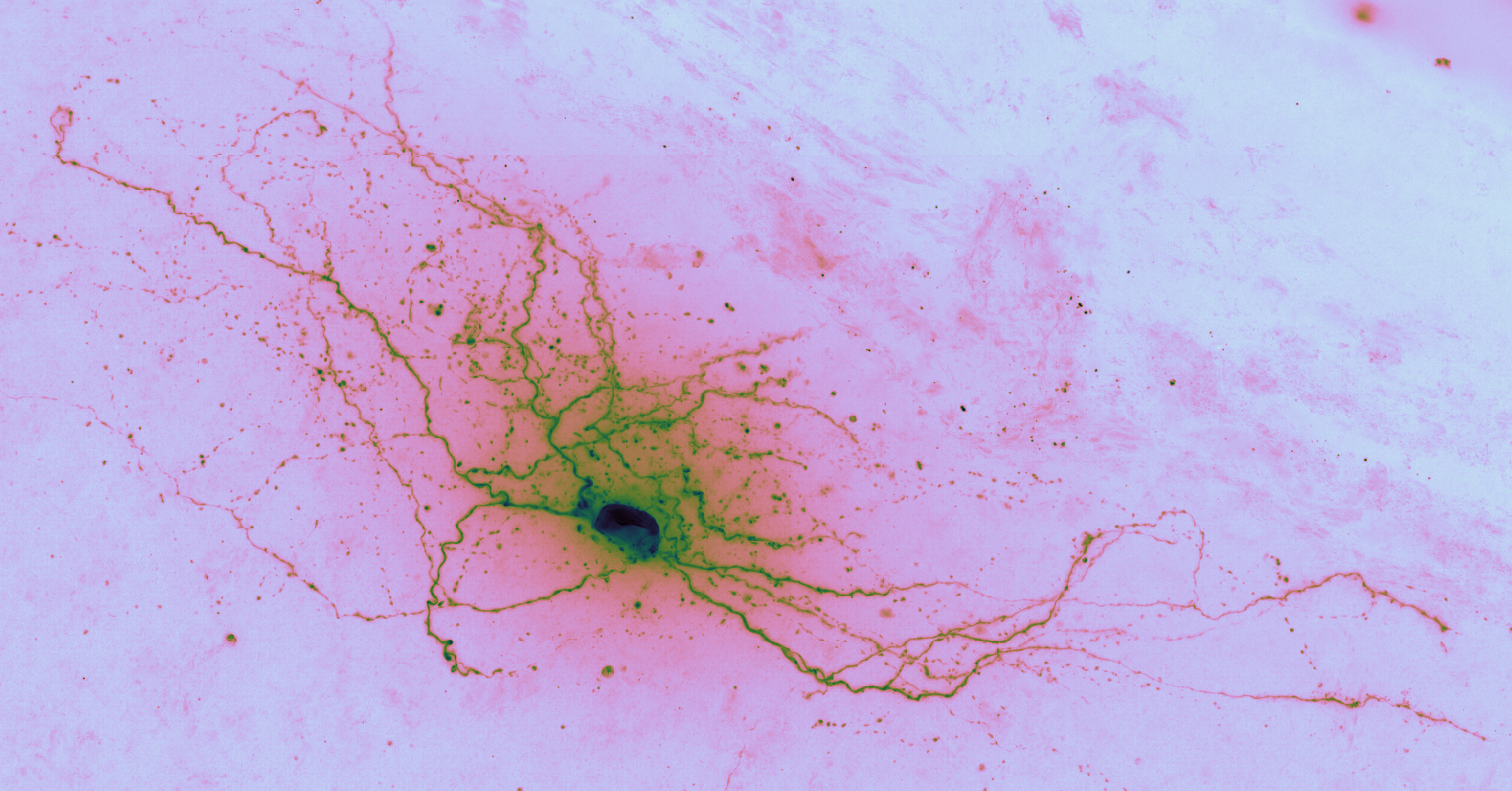
The first step with any dataset is exploratory data analysis and visualization. For a hands-on visual exploration of one cell’s data, see the notebook initialdatasetvisualization.cell_651806289.ipynb.
Target audience
This project uses the Brightfield Challenge dataset to train models for brightfield neuron skeletonization, which can subsequently be used in production. Both training and inference are computed on Colab, for free.
The target audience has two parts:
- Computer vision researchers building object recognizer (read: training)
- Lab scientists processing raw brightfield z-stacks into SWC files (read: inference)
The first audience is the computer vision software developers. This project provides Jupyter notebooks that perform data ETL, manifesting, and triaging. These notebooks perform set up and grunt data wrangling for using the dataset for training. Additionally, ample visualization tools are provided specific to the nature of data. These tools enables a developer to quickly get to the interesting part, with the tools to introspect the models and their output. For example, these notebooks:
- Access and download the dataset
- Process the data to generate SWC skeleton files
- Juxtapose new reconstruction SWCs alongside manual gold standard SWCs
The second audience is the sole researcher with a raw image stack off of a microscope desiring to produce a SWC, as well as other visualization. This person doesn’t want to train neural networks, they simply wish to upload their images and run software (with pre-trained ML models) that will make an SWC file and related renderings. This is the inference phase.
Background
The goal of auto-reconstruction is to automate neurite tracing, traditionally performed manually. The following image is taken from the challenge’s home page on alleninstitute.org. It illustrates the objective of this exercise: the input is grayscale microscope images of an individual neuron; the output is a 3D stick figure of the neuron.
In this image there are three examples, each in a separate column. The pairs of images consist of one grayscale camera image (a projection of the input) and one corresponding skeleton (the output) rendered in neon color on a dark field.
Brightfield modality
The majority of dendritic and axonal morphology reconstructions to date are based on bright-field microscopy (Halavi et al., 2012), due to its broad compatibility with histological staining methods… Moreover, the ability to enhance the signal intensity by counterstaining renders bright-field microscopy largely unsurpassed for reconstructions of whole axonal arbors up to the very thin (and typically faint) terminals. [Parekh and Ascoli, 2013]
In brightfield microscopy, a single neuron is pumped full of biocytin to stain its insides black, while the rest of the specimen’s brain tissue is chemically cleared to be translucent. In other words, there is a single dark/opaque foreground object – a biocytin stained neuron, the object of interest – which is imaged upon some essentially translucent background (or “field”). The field is bright (because of light shining through it) and the foreground is more opaque and so appears dark, ergo “brigthfield.”
For a quick-and-dirty overview of the actual wet bench rigmarole involved in generating the biocytin stained samples, see the 8 minute vidoe in Immunostaining of Biocytin-filled and Processed Sections for Neurochemical Markers (2016). There is also an 11 minute 2018 video on JoVE, walking through the nitty-gritty wet bench protocol for imaging neurons with biocytin, which demonstrates manual neuron tracing via Neurolucida.
For a backgrounder, see the 2007 article out of the Max Planck Institute, Transmitted light brightfield mosaic microscopy for three-dimensional tracing of single neuron morphology, or see PubMed for more on using biocytin to stain and trace neurons.
Reconstruction: an unsolved problem
Brightfield neuron reconsturction is an open image processing problem, and a rate limiting factor in brightfield microscopy. Currently, brightfield neuron reconstruction involves manual labor comprising many hours of manually tracing skeletons from the raw brightfield image stacks.
Brightfield microscopes are the simplest and most common type of microscope, so solving this problem could enable many labs to perform image analysis more quickly. This means that brightfield reconstruction software is more widely used, compared to other microscopy modalities. Brightfield is a relatively low tech microscopy modality that produces great results.
From the computer vision perspective, natural intelligence object recognition may be based on skeletons so this problem for neuroscience may feedback to the artificial intelligence community, specifically the visual object recongition folks. Recurrent networks are a promising candidate for this mode of microscopy data.
For a backgrounder, check out Neuronal Morphology Goes Digital: A Research Hub for Cellular and System Neuroscience Parekh & Ascoli (2013, in Neuron). Here’s an image from that paper illustrating the diversity of neuron morphologies across species:

Dataset analysis
In this project, Google Colab is used as a platform for reproducable research, specifically image analysis of biocytin stained neurons imaged via brightfield microscopy.
Platform: Google Colab
This project consists of Jupyter notebooks tuned up to run on Google Colab. The Colab service is Google’s free Jupter hosting service, packaged silimar to Google Sheets and Docs. An optional Nvidia GPU can be requested, useful for, say, GPU accelerated U-Net.
Jupyter notebooks are a popular medium for doing data science. Notebooks are a medium within which both computer programmers and neuroscientist are comfortable.
Colab is used to both perform analysis (e.g. generate projects and skeletons) and to publish results in pre-run notebooks. The curious can also re-run the notebooks to reproduce the results, or use the notebooks as workflow vignettes to spin off of.
Dataset specifics
The data is collected from a slice of brain sandwiched between a glass slide and a cover slip. The microscope’s field of view is much smaller than the slide so the slice is imaged tile by overlapping tile. The tiles are then stitched together to make a single 2D image, a virtual plate.
The third dimension of the image stack is created as the neuron is moved through the microscopes’s field of view by a motorized stage upon which the specimen slide is mounted. The stage drops about 5 micrometers at a step.
An artifact of this technique is that the images at the top and the bottom of the stack seem blurry, which is caused by the darkly stained neuron being out-of-focus – the so called bokeh effect, “the way the lens renders out-of-focus points of” darkness.
The specifics of how the data in the Challenge dataset was collected can be found in The Allen’s documentation, Allen Cell Types Database whitepaper, Cell Morphology And Histology, CellTypesMorphOverview.pdf:
Serial images (63X magnification) through biocytin-filled neurons…
Individual cells were imaged at higher resolution for the purpose of automated and manual reconstruction. Series of 2D images of single neurons were captured with a 63X objective lens (Zeiss Plan APOCHROMAT 63X/1.4 oil, 39.69x total magnification, and an n oil-immersion condenser 1.4 NA), using the Tile & Position and Z-stack ZEN 2012 SP2 software modules (Zeiss). The composite 2D tiled images with X-Y effective pixel size of 0.114 micron x 0.114 micron were acquired at an interval of 0.28 µm along the Z-axis.
Initial dataset exploration
So, what does this data actually look like? Here are two Jupyter notebooks that perform initial exploration and visualization of the challenge dataset:
- dataset_manifest.ipynb: file level overview of dataset; creates
specimens_manifest.json - initial-dataset-visualization.ipynb: visual deep dive of a single specimen’s imagery; uses
specimens_manifest.json