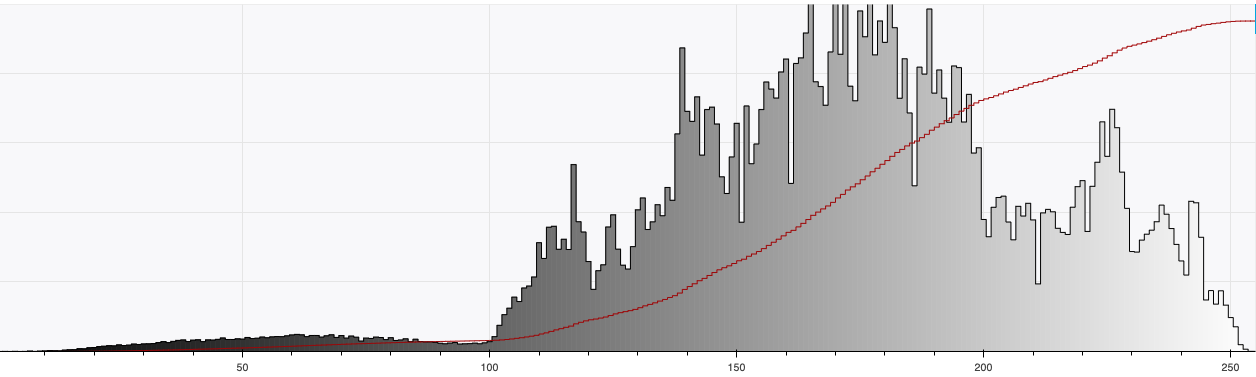
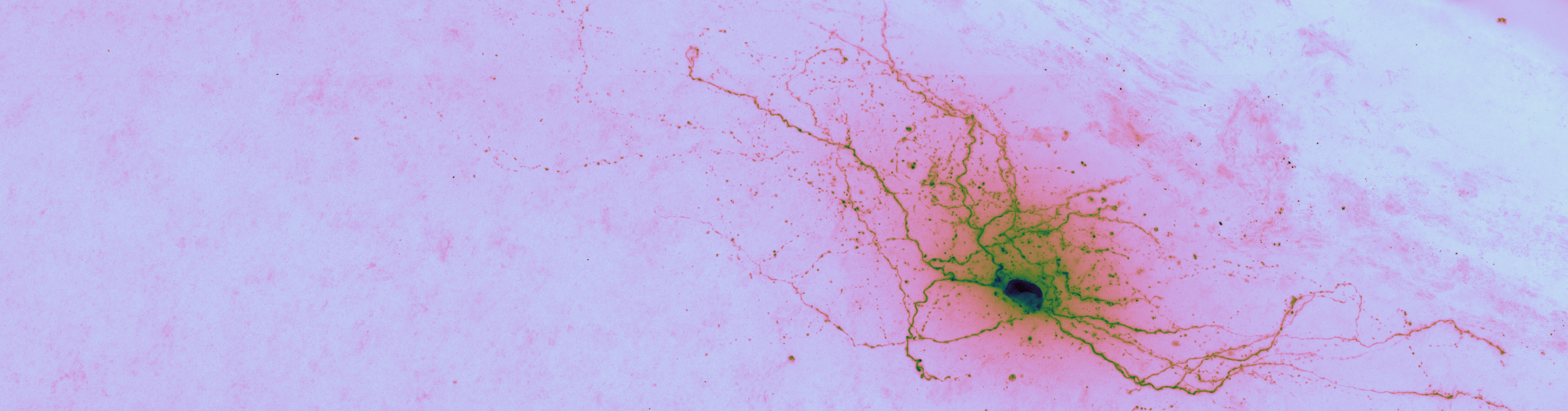
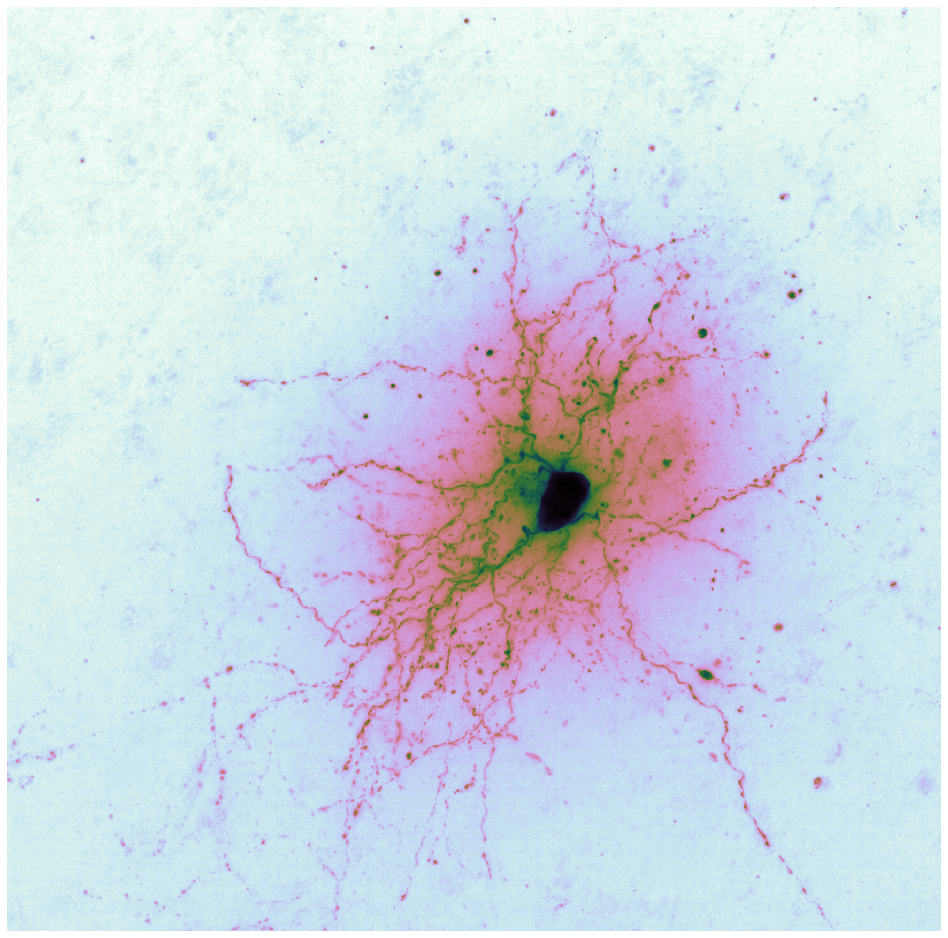
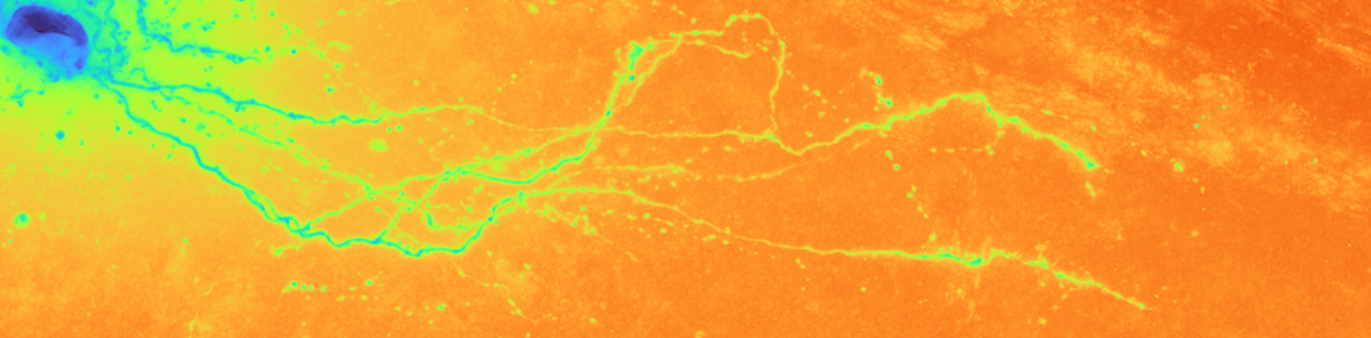
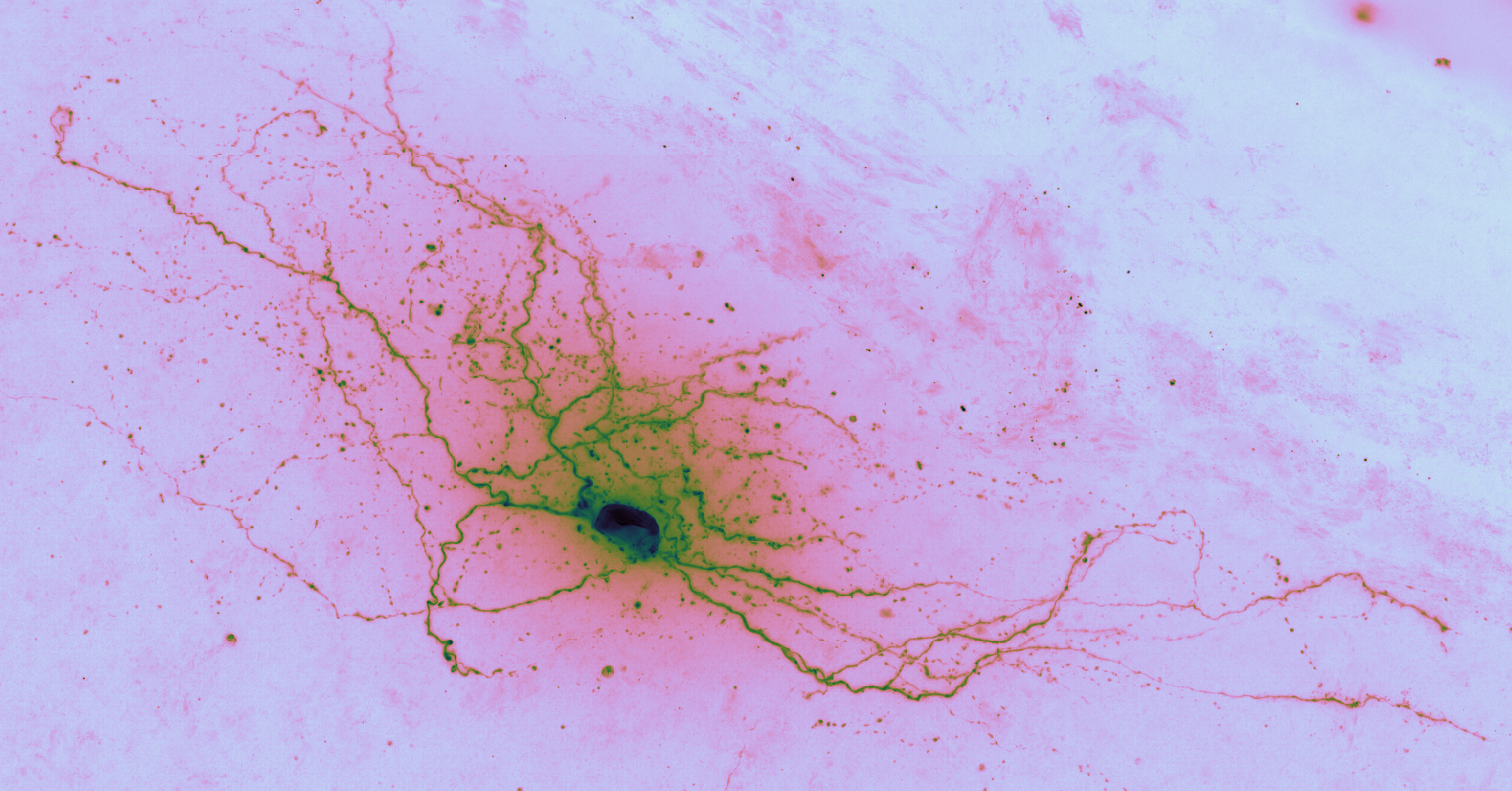
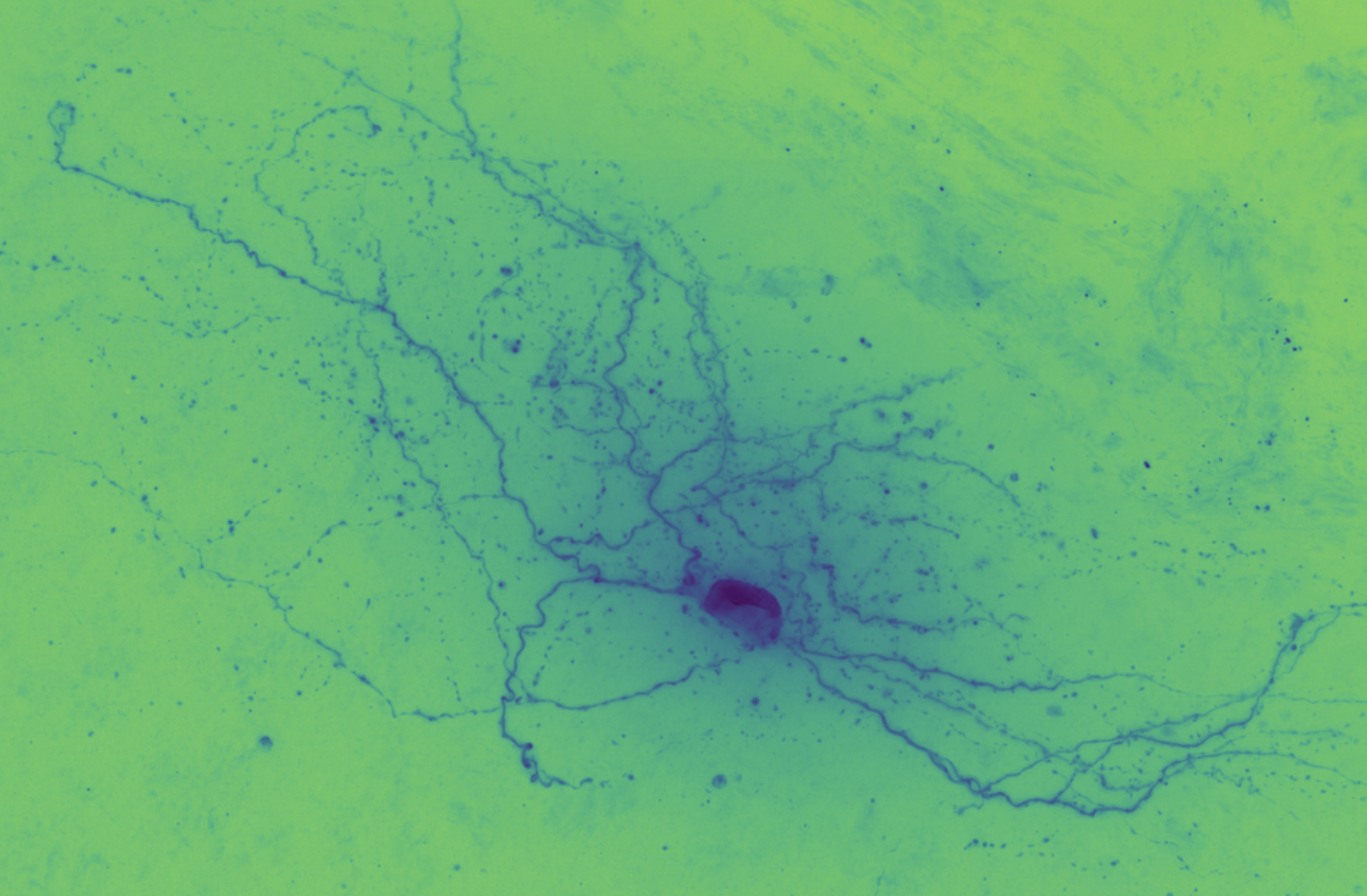
Brightfield data is single channel. It is not color data, which is triple band data (Red, Green, Blue or such). Therefore there is the opportunity to colorize the images according to some map. Pseudocoloring can be seen as some of the simpliest form of image processing for brightfield microscopy. As can be seen in this page’s header imageset, different colormaps bring out different features of an image.
The whole goal of this project is to make tools which make it easier to gain insights from the raw images off the microscope. Colormapping is about as basic as it gets but it should be addressed in the current Jupyter-based context of Python on the server and JavaScript on the client.
This code is being written in 2020 so let’s color these micrographs like it’s 2020, with decent color maps that work well with brihtfield images. In particular 2019 saw the publication of the Turbo colormap by Google AI which looks good – loud but effective at separating the foreground from backfield (e.g., it’s the reddish image at the top of this page).
Further, this codebase is Jupyter-based. As used in this project, that means Python on the Jupyter server and JavaScript in the browser. It would be nice to use the same colormaps in images produced by both Python and JavaScript. As such, this project has been built out using a few colormaps that seem to work well.
Jet is the default “rainbow” colormap in Python-land. Jet is a lame implementation. There are other rainbow colormaps, say, Turbo.
The following juxtaposition illustrates Jet’s flaws (images are from Google’s post).
So, it’s not a complete win but Turbo is easier to look at than Jet and brings out more features that Jet.
Turbo is not yet distributed with Python’s Matplotlib. But adding a new colormap for use by Matplotlib is only a handful of lines of code. At its core a Matplotlib colormap is just an array of 256 values, each a color (a RGB triplet) which Matplotlib wants normalized into a float between 0 and 1, rather than an 8-bit 0 to 255 range which is what HTML expects.
turbo_colormap_data = [[0.18995,0.07176,0.23217],[0.19483,0.08339,0.26149],[0.19956,0.09498,0.29024],[0.20415,0.10652,0.31844],[0.20860,0.11802,0.34607],[0.21291,0.12947,0.37314],[0.21708,0.14087,0.39964],[0.22111,0.15223,0.42558],[0.22500,0.16354,0.45096],[0.22875,0.17481,0.47578],[0.23236,0.18603,0.50004],[0.23582,0.19720,0.52373],[0.23915,0.20833,0.54686],[0.24234,0.21941,0.56942],[0.24539,0.23044,0.59142],[0.24830,0.24143,0.61286],[0.25107,0.25237,0.63374],[0.25369,0.26327,0.65406],[0.25618,0.27412,0.67381],[0.25853,0.28492,0.69300],[0.26074,0.29568,0.71162],[0.26280,0.30639,0.72968],[0.26473,0.31706,0.74718],[0.26652,0.32768,0.76412],[0.26816,0.33825,0.78050],[0.26967,0.34878,0.79631],[0.27103,0.35926,0.81156],[0.27226,0.36970,0.82624],[0.27334,0.38008,0.84037],[0.27429,0.39043,0.85393],[0.27509,0.40072,0.86692],[0.27576,0.41097,0.87936],[0.27628,0.42118,0.89123],[0.27667,0.43134,0.90254],[0.27691,0.44145,0.91328],[0.27701,0.45152,0.92347],[0.27698,0.46153,0.93309],[0.27680,0.47151,0.94214],[0.27648,0.48144,0.95064],[0.27603,0.49132,0.95857],[0.27543,0.50115,0.96594],[0.27469,0.51094,0.97275],[0.27381,0.52069,0.97899],[0.27273,0.53040,0.98461],[0.27106,0.54015,0.98930],[0.26878,0.54995,0.99303],[0.26592,0.55979,0.99583],[0.26252,0.56967,0.99773],[0.25862,0.57958,0.99876],[0.25425,0.58950,0.99896],[0.24946,0.59943,0.99835],[0.24427,0.60937,0.99697],[0.23874,0.61931,0.99485],[0.23288,0.62923,0.99202],[0.22676,0.63913,0.98851],[0.22039,0.64901,0.98436],[0.21382,0.65886,0.97959],[0.20708,0.66866,0.97423],[0.20021,0.67842,0.96833],[0.19326,0.68812,0.96190],[0.18625,0.69775,0.95498],[0.17923,0.70732,0.94761],[0.17223,0.71680,0.93981],[0.16529,0.72620,0.93161],[0.15844,0.73551,0.92305],[0.15173,0.74472,0.91416],[0.14519,0.75381,0.90496],[0.13886,0.76279,0.89550],[0.13278,0.77165,0.88580],[0.12698,0.78037,0.87590],[0.12151,0.78896,0.86581],[0.11639,0.79740,0.85559],[0.11167,0.80569,0.84525],[0.10738,0.81381,0.83484],[0.10357,0.82177,0.82437],[0.10026,0.82955,0.81389],[0.09750,0.83714,0.80342],[0.09532,0.84455,0.79299],[0.09377,0.85175,0.78264],[0.09287,0.85875,0.77240],[0.09267,0.86554,0.76230],[0.09320,0.87211,0.75237],[0.09451,0.87844,0.74265],[0.09662,0.88454,0.73316],[0.09958,0.89040,0.72393],[0.10342,0.89600,0.71500],[0.10815,0.90142,0.70599],[0.11374,0.90673,0.69651],[0.12014,0.91193,0.68660],[0.12733,0.91701,0.67627],[0.13526,0.92197,0.66556],[0.14391,0.92680,0.65448],[0.15323,0.93151,0.64308],[0.16319,0.93609,0.63137],[0.17377,0.94053,0.61938],[0.18491,0.94484,0.60713],[0.19659,0.94901,0.59466],[0.20877,0.95304,0.58199],[0.22142,0.95692,0.56914],[0.23449,0.96065,0.55614],[0.24797,0.96423,0.54303],[0.26180,0.96765,0.52981],[0.27597,0.97092,0.51653],[0.29042,0.97403,0.50321],[0.30513,0.97697,0.48987],[0.32006,0.97974,0.47654],[0.33517,0.98234,0.46325],[0.35043,0.98477,0.45002],[0.36581,0.98702,0.43688],[0.38127,0.98909,0.42386],[0.39678,0.99098,0.41098],[0.41229,0.99268,0.39826],[0.42778,0.99419,0.38575],[0.44321,0.99551,0.37345],[0.45854,0.99663,0.36140],[0.47375,0.99755,0.34963],[0.48879,0.99828,0.33816],[0.50362,0.99879,0.32701],[0.51822,0.99910,0.31622],[0.53255,0.99919,0.30581],[0.54658,0.99907,0.29581],[0.56026,0.99873,0.28623],[0.57357,0.99817,0.27712],[0.58646,0.99739,0.26849],[0.59891,0.99638,0.26038],[0.61088,0.99514,0.25280],[0.62233,0.99366,0.24579],[0.63323,0.99195,0.23937],[0.64362,0.98999,0.23356],[0.65394,0.98775,0.22835],[0.66428,0.98524,0.22370],[0.67462,0.98246,0.21960],[0.68494,0.97941,0.21602],[0.69525,0.97610,0.21294],[0.70553,0.97255,0.21032],[0.71577,0.96875,0.20815],[0.72596,0.96470,0.20640],[0.73610,0.96043,0.20504],[0.74617,0.95593,0.20406],[0.75617,0.95121,0.20343],[0.76608,0.94627,0.20311],[0.77591,0.94113,0.20310],[0.78563,0.93579,0.20336],[0.79524,0.93025,0.20386],[0.80473,0.92452,0.20459],[0.81410,0.91861,0.20552],[0.82333,0.91253,0.20663],[0.83241,0.90627,0.20788],[0.84133,0.89986,0.20926],[0.85010,0.89328,0.21074],[0.85868,0.88655,0.21230],[0.86709,0.87968,0.21391],[0.87530,0.87267,0.21555],[0.88331,0.86553,0.21719],[0.89112,0.85826,0.21880],[0.89870,0.85087,0.22038],[0.90605,0.84337,0.22188],[0.91317,0.83576,0.22328],[0.92004,0.82806,0.22456],[0.92666,0.82025,0.22570],[0.93301,0.81236,0.22667],[0.93909,0.80439,0.22744],[0.94489,0.79634,0.22800],[0.95039,0.78823,0.22831],[0.95560,0.78005,0.22836],[0.96049,0.77181,0.22811],[0.96507,0.76352,0.22754],[0.96931,0.75519,0.22663],[0.97323,0.74682,0.22536],[0.97679,0.73842,0.22369],[0.98000,0.73000,0.22161],[0.98289,0.72140,0.21918],[0.98549,0.71250,0.21650],[0.98781,0.70330,0.21358],[0.98986,0.69382,0.21043],[0.99163,0.68408,0.20706],[0.99314,0.67408,0.20348],[0.99438,0.66386,0.19971],[0.99535,0.65341,0.19577],[0.99607,0.64277,0.19165],[0.99654,0.63193,0.18738],[0.99675,0.62093,0.18297],[0.99672,0.60977,0.17842],[0.99644,0.59846,0.17376],[0.99593,0.58703,0.16899],[0.99517,0.57549,0.16412],[0.99419,0.56386,0.15918],[0.99297,0.55214,0.15417],[0.99153,0.54036,0.14910],[0.98987,0.52854,0.14398],[0.98799,0.51667,0.13883],[0.98590,0.50479,0.13367],[0.98360,0.49291,0.12849],[0.98108,0.48104,0.12332],[0.97837,0.46920,0.11817],[0.97545,0.45740,0.11305],[0.97234,0.44565,0.10797],[0.96904,0.43399,0.10294],[0.96555,0.42241,0.09798],[0.96187,0.41093,0.09310],[0.95801,0.39958,0.08831],[0.95398,0.38836,0.08362],[0.94977,0.37729,0.07905],[0.94538,0.36638,0.07461],[0.94084,0.35566,0.07031],[0.93612,0.34513,0.06616],[0.93125,0.33482,0.06218],[0.92623,0.32473,0.05837],[0.92105,0.31489,0.05475],[0.91572,0.30530,0.05134],[0.91024,0.29599,0.04814],[0.90463,0.28696,0.04516],[0.89888,0.27824,0.04243],[0.89298,0.26981,0.03993],[0.88691,0.26152,0.03753],[0.88066,0.25334,0.03521],[0.87422,0.24526,0.03297],[0.86760,0.23730,0.03082],[0.86079,0.22945,0.02875],[0.85380,0.22170,0.02677],[0.84662,0.21407,0.02487],[0.83926,0.20654,0.02305],[0.83172,0.19912,0.02131],[0.82399,0.19182,0.01966],[0.81608,0.18462,0.01809],[0.80799,0.17753,0.01660],[0.79971,0.17055,0.01520],[0.79125,0.16368,0.01387],[0.78260,0.15693,0.01264],[0.77377,0.15028,0.01148],[0.76476,0.14374,0.01041],[0.75556,0.13731,0.00942],[0.74617,0.13098,0.00851],[0.73661,0.12477,0.00769],[0.72686,0.11867,0.00695],[0.71692,0.11268,0.00629],[0.70680,0.10680,0.00571],[0.69650,0.10102,0.00522],[0.68602,0.09536,0.00481],[0.67535,0.08980,0.00449],[0.66449,0.08436,0.00424],[0.65345,0.07902,0.00408],[0.64223,0.07380,0.00401],[0.63082,0.06868,0.00401],[0.61923,0.06367,0.00410],[0.60746,0.05878,0.00427],[0.59550,0.05399,0.00453],[0.58336,0.04931,0.00486],[0.57103,0.04474,0.00529],[0.55852,0.04028,0.00579],[0.54583,0.03593,0.00638],[0.53295,0.03169,0.00705],[0.51989,0.02756,0.00780],[0.50664,0.02354,0.00863],[0.49321,0.01963,0.00955],[0.47960,0.01583,0.01055]]
turbo_cmap=ListedColormap(turbo_colormap_data)
Viridis is the default colormap for Matplotlib. It doesn’t suck.
Cubehelix is a modern colormap that works well with brightfield because the top end of the colormap fades out to white, much like the bright field in brightfield.
“Cubehelix was created to vary smoothly in both lightness and hue, but appears to have a small hump in the green hue area.” Seems to be a 2011 thing out of the astronomy world.
In the end, Turbo seems like a good default.